Newsletter Signup - Under Article / In Page
"*" indicates required fields
Nick McCooke shares the story behind the invention of next-generation sequencing at Solexa and his latest adventure in the sequencing world with the company DNA Electronics.
Nick McCooke led the pioneer team at Solexa that invented next-generation sequencing, a technology to read DNA at high speed that is nowadays used worldwide and has laid the foundation for precision medicine.
Solexa was acquired by Illumina in 2006 for what amounted to around €500M back then. Today, Illumina controls the majority of the global next-generation sequencing market with the technology from Solexa.
Since then, McCooke has been involved with the sequencing field working with companies such as DNA Electronics, which is part of a wave of new technologies to make sequencing even faster and cheaper. Back when working at Solexa, McCooke’s ambitious goal was the $1,000 genome. Today the goal is $100.
I spoke with McCooke to hear first-hand about his journey with both Solexa and DNA Electronics, the ups and downs he faced on the way, and the excitement that inevitably comes from making a breakthrough after lots of hard work.
Let’s start with Solexa. How did you get involved with it?
I joined Solexa in 2000, which just shows you how long ago next-generation sequencing started. I had been working in the US, and I had sold the company that I was working for. I came back to the UK looking for the next exciting thing to do and I ended up setting the company up as the founding CEO.
Now, next-generation sequencing is kind of part of the furniture, it feels as if it’s always been around. But back then, of course, it didn’t feel like that. We were in the middle of the Human Genome Project, there were factories full of the old 3730 Sanger sequencers, and no one had thought beyond that.

The initial idea came from a couple of guys at Cambridge University. The concept of next-generation sequencing is basically to massively parallelize the sequencing process. The ABI 3730 sequencer was processing 96 strands of DNA in parallel – highly accurately, actually, but only 96 lanes. Our first instrument had something like 40 million interrogation sites on the array. We realized it could impact speed and cost by several orders of magnitude.
I really wanted to inspire people – hopefully investors, anyway – and give the company an ambitious goal to head towards. So, I came up with the idea of the ‘thousand-dollar genome.’ We knew we couldn’t achieve that immediately, but our calculations showed it was possible.
It was hard work, and some things didn’t work at first. The original idea was to basically chop up DNA and put single molecules on an array slide. But it was a complete dead end. We managed to find an alternative, which became the bridge amplification technology in the Illumina system. I negotiated to buy that technology from a Swiss company called Manteia, and that really saved the day. If we hadn’t been able to acquire that technology, the project would have failed.
There were lots of ups and downs, but by early 2005, we’d got it working and we sequenced the very first small genome, from the virus phiX-74. The company went onto the NASDAQ roughly around that time, and then about eighteen months later it was acquired by Illumina.
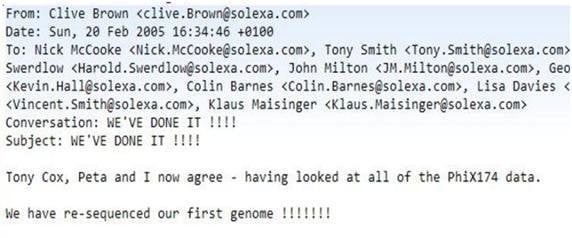
With such a success in your hands, why did you leave before the acquisition?
Well, because the company moved to the US, and my family and I weren’t in a position to go back there. And we’d already done all the difficult stuff.
After I left Solexa, I did a number of other things – life’s too short to spend it doing one thing. But I came back to sequencing with DNA Electronics. I hadn’t really been looking around too much about what was happening in sequencing, and I found not an awful lot had changed. Certainly, the performance of Illumina and other sequencers had improved. But they were still doing pretty much the same thing.
To come into the market now, you have to do something very different. There’s no point in competing with Illumina unless you’ve got something which is a lot better or you’re doing something very, very different.
Tell me more about this new technology being developed at DNA Electronics.
DNA Electronics works on semiconductor sequencing. This is a technology invented by the founder, Chris Toumazou, who is a Professor at Imperial College London. It’s based on using standard semiconductor chips to detect and measure sequencing reactions.
Basically, when you add a nucleotide to a template – which is the classic way of doing sequencing by synthesis – a hydrogen ion is released. On these microchips, there’s an array of lots and lots of ‘ion-sensitive field effect transistors’ that detect the release of those hydrogen ions.
This technology has been implemented by Ion Torrent, now part of Thermo Fisher. They are a licensee, so they pay a royalty when they sell their semiconductor sequencing chips. But unlike Thermo Fisher, which focuses on research applications, DNA Electronics is solely focused on applying next-generation sequencing to clinical diagnostics.
What advantages could this new technology bring to diagnostics?
If you want to use conventional sequencing in diagnostics, the workflow currently requires quite a lot of preparation outside of the sequencer. And then there is the bioinformatics afterwards. So, it’s a long and complicated process that can only be done in a specialized laboratory with skilled operators. It takes days. And it’s not a closed system, so there’s risk of contamination.
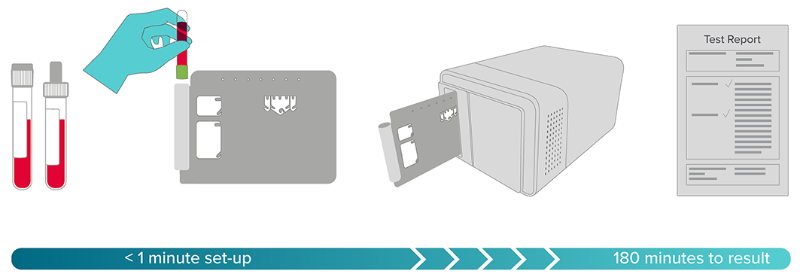
If you look at PCR platforms like Cepheid, they have been designed to be simple to the user, and we don’t see why sequencing should be any different. Our basic concept is putting the sample into a cartridge, the cartridge into an instrument, press a button, and two to four hours later you get your answer.
Our approach can take place pretty much anywhere, and doesn’t require a high level of skill. Because the technology is closer to the patient, we can get results to the doctors who are treating the patient much more quickly, within the hospital. Whereas if you use conventional sequencing, that sample may have to travel tens or possibly hundreds of miles to get to the right lab. Then it probably joins a queue before the sequencing gets done. And then the results have to be sent back to the hospital.
What’s your view on other NGS companies like Oxford Nanopore?
Incidentally, I know quite a lot of the people at Oxford Nanopore because they were part of my team at Solexa, where we pioneered the next generation of sequencing. It was a fantastic team and I think we were part of something very, very special. Some are still in Illumina, and some have gone to Oxford Nanopore or other sequencing companies. We keep in touch, and there’s that sort of ‘band of brothers and sisters’ feel to it.
Back to Oxford Nanopore, the company is making a very small sequencer, but actually, in a hospital laboratory that doesn’t make a lot of difference. Oxford Nanopore is still in the research market, where it sells a sequencer that can be used in many different ways but isn’t focused on a particular application.
I think it’s very important to look at the technology you have, and understand where its real strengths are, and what markets and opportunities it’s best suited to. In our case, we clearly saw that semiconductor sequencing was best applied in the clinical diagnostic area.
What are your plans for the coming years?
I’ve hugely enjoyed getting back to sequencing, and I think there’s a lot more to achieve. Now it’s not the old goal of how quickly we can sequence the human genome. Now it’s, “how can we use sequencing to improve human health?” That’s where I think sequencing is headed now.
One of the advantages of sequencing in this field is that we can really go into depth in terms of antimicrobial resistance markers. DNA Electronics is launching a test that will be used to rapidly identify the infectious agents that can lead to sepsis – a market where speed of results is critical in saving lives and containing costs. And the US Government organization BARDA is supporting the development of two tests for antimicrobial resistance and flu virus.
The company also has a particular interest in cancer. There’s a lot of talk about liquid biopsy, pulling out human DNA from blood, and the platform is highly suited to that application.

Before you go, I’m just curious: do you think there’s a reason most sequencing technologies are developed in the UK?
It’s interesting, isn’t it? Sanger, Solexa, DNA Electronics, Oxford Nanopore… The inventions were independent, but I think once you have a skill base developed within a country, it does mean you have a critical mass of people who have worked in the field. And, of course, they can redeploy in other companies. But I don’t really know the answer. Something in the water we drink, perhaps?
This article was originally published on October 2017 and has been republished with minor edits.
Images via Shutterstock; ThermoFisher; DNA Electronics






