Newsletter Signup - Under Article / In Page
"*" indicates required fields
With the rise of a whole new field of research around the microbiome, what role are new technologies to identify microorganisms playing in the future of healthcare?
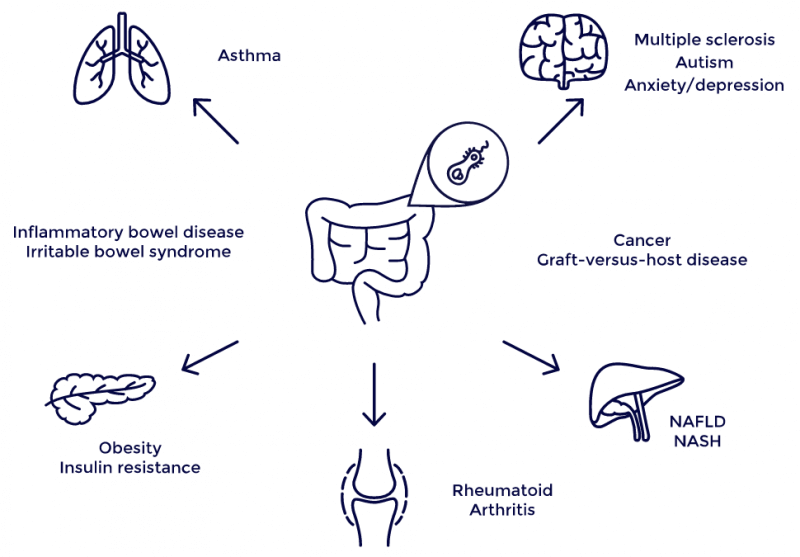
Traditionally, microbes have been seen solely as a threat. But in the last few years, microbiome research has started to shift this view. Today, we know certain microorganisms are actually essential for our health, while others are responsible for infections and can be instrumental in causing all sorts of diseases, including inflammatory bowel disease or type 2 diabetes. They can even affect medical treatments; some microorganisms are known to render cancer drugs ineffective, whereas others are actually necessary to make these drugs work.
This means that broad-range antimicrobials can sometimes cause more problems in our health than they fix, because they cannot discriminate between ‘good’ and ‘bad’ bacteria. There is now a pressing need to be able to accurately identify microbes in order to study their contribution to disease and design treatments that only attack pathogenic bacteria.
In hospitals, the traditional method to identify microorganisms is through culture tests, a slow method that requires waiting for several days while bacteria grow in the lab. In addition, it is estimated that 90% of bacteria can’t be detected using this method.
The advent of next-generation sequencing has started to challenge culture tests. These techniques can sequence all the DNA present in a sample, including that of bacteria that may have been killed by preventive treatment with broad-range antimicrobials.
Another common technique is qPCR. It works by amplifying an already known DNA sequence and quantifying how much of it is found in the sample. Since it is targeted against a specific sequence, it is particularly useful to obtain results faster when you already know what you’re looking for.
Sequencing and qPCR can be complementary; the former can be used to identify relevant disease biomarkers that then are translated into a qPCR test. That’s what Microbiome Diagnostics Partners, a subsidiary of the French microbiome therapeutics company Enterome, is doing.
“We have been looking at the evolution of the gut microbiome composition over time in Crohn’s disease,” Pierre Belichard, CEO of Enterome, told me. “We have identified bacteria that are always downregulated or upregulated when the disease is active, and developed a test that can replace colonoscopy.”
This microbiome test, which Belichard expects to see in the market in 2019, is designed to be a companion for a microbiome-targeting treatment being developed by Enterome and Takeda against Crohn’s disease. Soon, it could be followed by other microbiome diagnostics and treatment combos for liver disease and even cancer.
But these techniques still pose some limitations. While diagnosing chronic diseases is important, other more acute conditions require a faster response than that of conventional technologies.
Oxford Nanopore Technologies has pioneered a new sequencing technology that can read DNA much faster and at a lower price using a pocket-sized device called MinION. The company is testing the device in microbiome diagnostic applications, including the detection of antibiotic resistance genes, to help determine the best treatment.
UK-based DNA Electronics is developing a similarly fast technology, in its case fully focused on diagnostic applications. “Our basic concept is putting the sample into a cartridge, the cartridge into an instrument, press a button, and two to four hours later you get your answer,” CEO Nick McCooke told me.
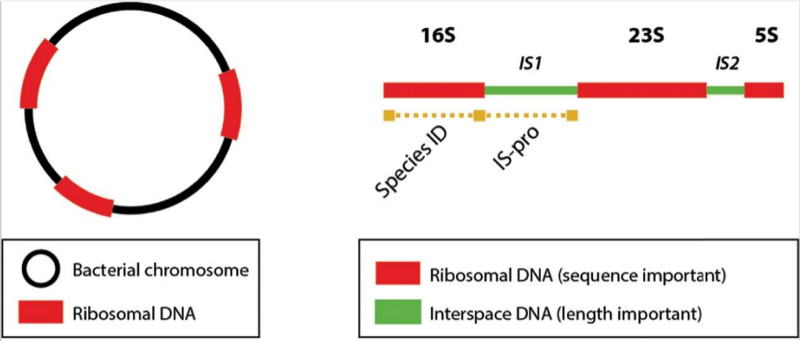
A small company in the Netherlands called IS Diagnostics is going after a whole new approach to improve microbiology diagnostics. The company targets a specific location of the bacterial DNA called the IS region, which is located just after the 16S region commonly used to identify the species of a bacterium based on its sequence. Rather than a distinct sequence, the IS region has a specific length for each species, which is easier to detect and reduces the time to make a diagnosis down to 4-5 hours.
The technique also includes a color-coding system to classify each broader group of bacteria to increase the resolution at which different microbes can be differentiated.
“It’s a universal technology to rapidly analyze all bacteria in a sample,” explains CEO Dries Budding. The technique is already in use for general analysis of bacteria for infectious diseases in hospitals, including the VU University Medical Center in Amsterdam, from which IS Diagnostics is a spin-off.
IS Diagnostics has launched a microbiome diagnostics kit, only for research so far, that has been validated for monitoring inflammatory bowel disease. The company is also working on monitoring specific strains of C. difficile species, which has become an urgent need according to the CDC.
“Microbiology is traditionally retrospective,” said Budding. “We aim for it to become predictive instead.” Indeed, with the rapid advances of treatments focused on microbes, new diagnostic technologies will play a key role in the way we treat all sorts of diseases, ranging from infections to cancer, and ultimately save lives.
This feature was originally published on 12.10.17. It has since been updated by the Labiotech editorial team to reflect the latest developments in the field.
Images via joker1991, Lutanski /Shutterstock; 4D pharma; J Clin Microbiol. 2016; 54(4): 934–943.